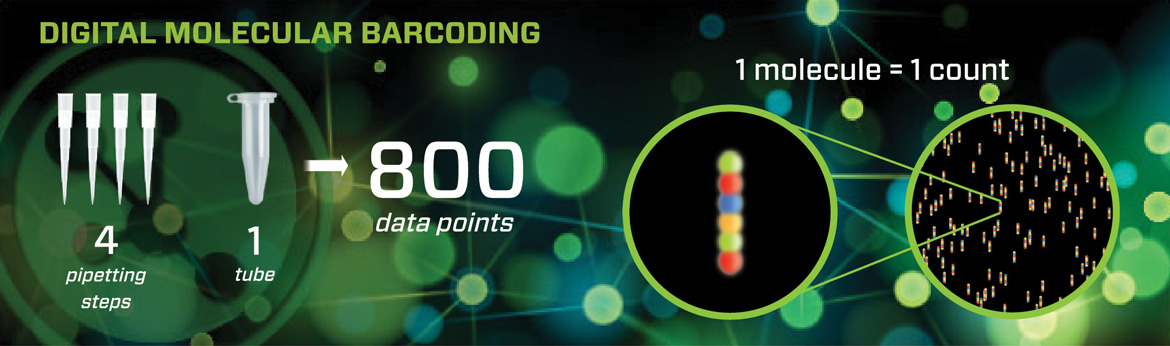
Nanostring's patented technology used in the nCounter Analysis System is a true digital detection technology capable of highly multiplexed, direct profiling of individual molecules in a single reaction without amplification. Barcoded probes hybridize directly to a target molecule in solution. The Reporter Probe carries the barcode, and the Capture Probe allows the complex to be immobilized for data collection.
Unparalleled Performance in Digital Gene Expression
nCounter Gene Expression CodeSets offer a cost-effective way to analyze the expression levels of up to 800 genes simultaneously, with precision superior to qPCR. The system can directly assay tissue and blood lysates as well as Formalin-Fixed Paraffin-Embedded (FFPE) extracts in a simple workflow that requires only 15 minutes of total hands-on time per reaction.
Product Highlights
 Analyze up to 800 genes simultaneously
Analyze up to 800 genes simultaneously
 No RT, no enzymes, no amplification*
No RT, no enzymes, no amplification*
*Single Cell analysis requires amplification prior to analysis on the nCounter System.
 Direct input of multiple sample types including total RNA, cell lysate, FFPE samples, and whole-blood lysate
Direct input of multiple sample types including total RNA, cell lysate, FFPE samples, and whole-blood lysate
 Single molecule digital counting minimizes data noise
Single molecule digital counting minimizes data noise
Data You Can Count On
The nCounter Gene Expression Assay is designed to provide an ultra-sensitive, reproducible and highly multiplexed method for gene expression profiling across all levels of biological expression. This assay provides a method for direct detection of mRNAs with molecular barcodes called nCounter Reporter Probes without the use of reverse transcription or amplification.
Sensitivity, Reproducibility and Dynamic Range
Gene counts from two technical replicates plotted against one another demonstrate the assay reproducibility over a wide dynamic range (10 - 50,000 counts). One total RNA sample was split into two separate hybridization reactions and processed independently on the nCounter Analysis System. In this experiment, 75 counts is equal to a concentration of approximately 1 copy per cell. These data illustrate the high level of sensitivity and precision of the assay even at very low levels of expression.
Nanostring Featured on Nature Biotechnology (March 2008)

"Direct Multiplexed Measurement of Gene Expression with Color-coded Probe Pairs" - by Geiss GK, et al. -
The paper entitled Direct Multiplexed Measurement of Gene Expression with Color-Coded Probe Pairs describes the performance of the nCounter Analysis System, which captures and counts individual mRNA transcripts by a novel molecular bar-coding technology. The new platform was compared to microarrays, TaqMan® PCR, and SYBR®Green real-time PCR and results demonstrated that the nCounter system is more sensitive than microarrays and is similar in sensitivity to real-time PCR. The paper describes experiments using nCounter CodeSets to profile the expression of 509 human genes in a single multiplexed assay, analysis of a gene set that overlaps with the MAQC gene expression study and quantitation of transcript levels for 21 sea urchin genes across 7 embryo samples. Results demonstrated a remarkable correlation in the pattern of gene expression between NanoString technology and real-time PCR at all transcript levels. The paper was a result of a collaboration between scientists at NanoString Technologies, Dr. Roger Bumgarner at the University of Washington, Dr. Eric Davidson at the California Institute of Technology, and Dr. Leroy Hood at the Institute of Systems Biology.
Abstract
We describe a technology, the NanoString nCounter gene expression system, which captures and counts individual mRNA transcripts. Advantages over existing platforms include direct measurement of mRNA expression levels without enzymatic reactions or bias, sensitivity coupled with high multiplex capability, and digital readout...
Complete Workflow from start to end
 |
 |
 |
| Pre-build panels for gene expression, miRNA, miRGE, CNV, fusion genes and protein OR custom panels. | Automated past-hybridization processing on nCounter systems. | nSolver utilizes a variety of appropriate normalization methodologies in all samples to provide accurate analysis. |